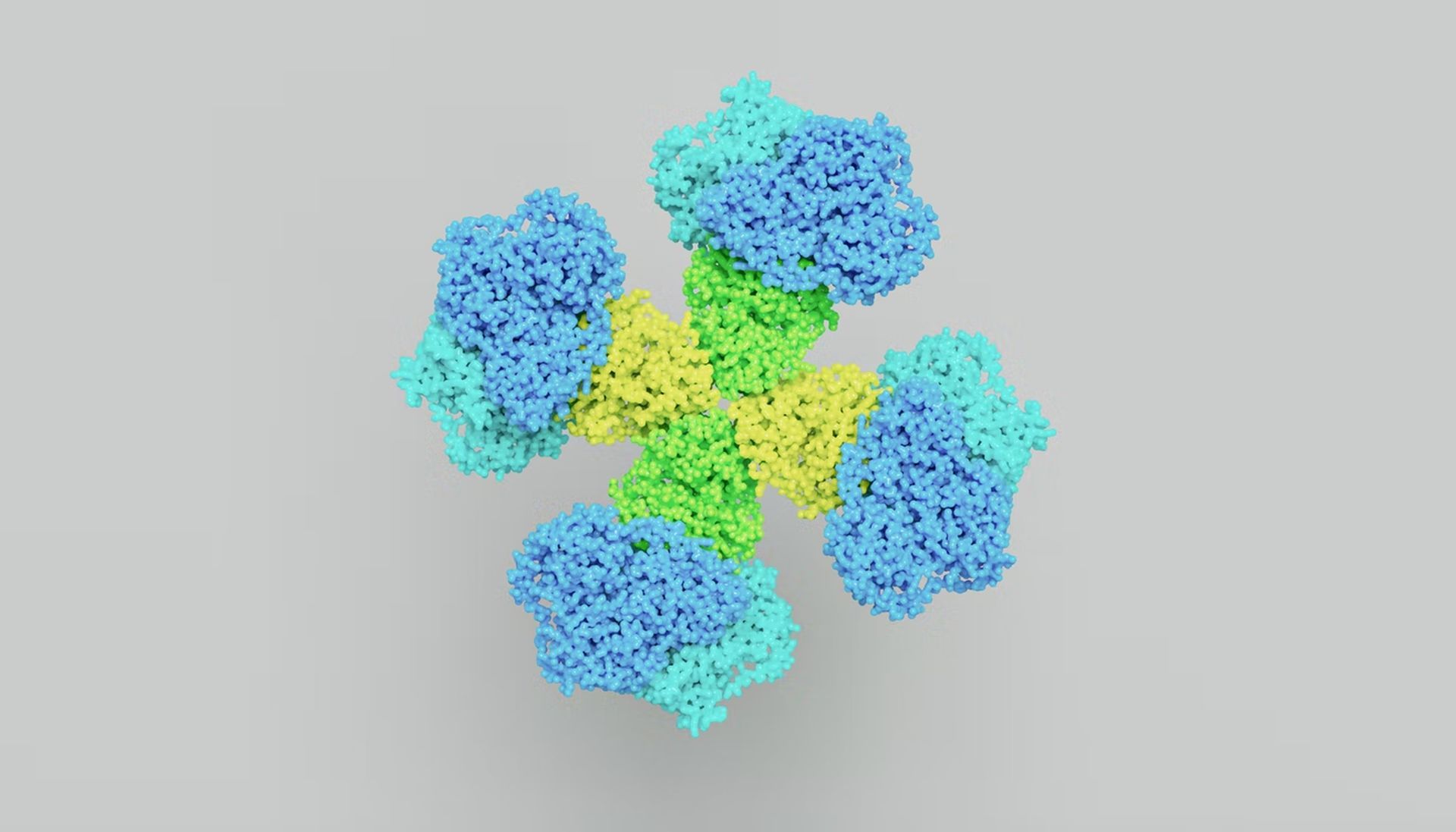
DeepMind revealed that the structures of all proteins known to science, more than 200 million proteins, have been solved by its AlphaFold AI.
Google’s DeepMind AI’s prediction of the 3D structures of all proteins known to science represents a major advance that could enhance our understanding of rare genetic diseases and enable the development of novel vaccinations and treatments.
AlphaFold learns to decode the remaining protein folding structures
The building blocks of life, proteins serve various functions in the body as structural components, transport molecules, and enzymes, which operate as functional catalysts for chemical reactions.
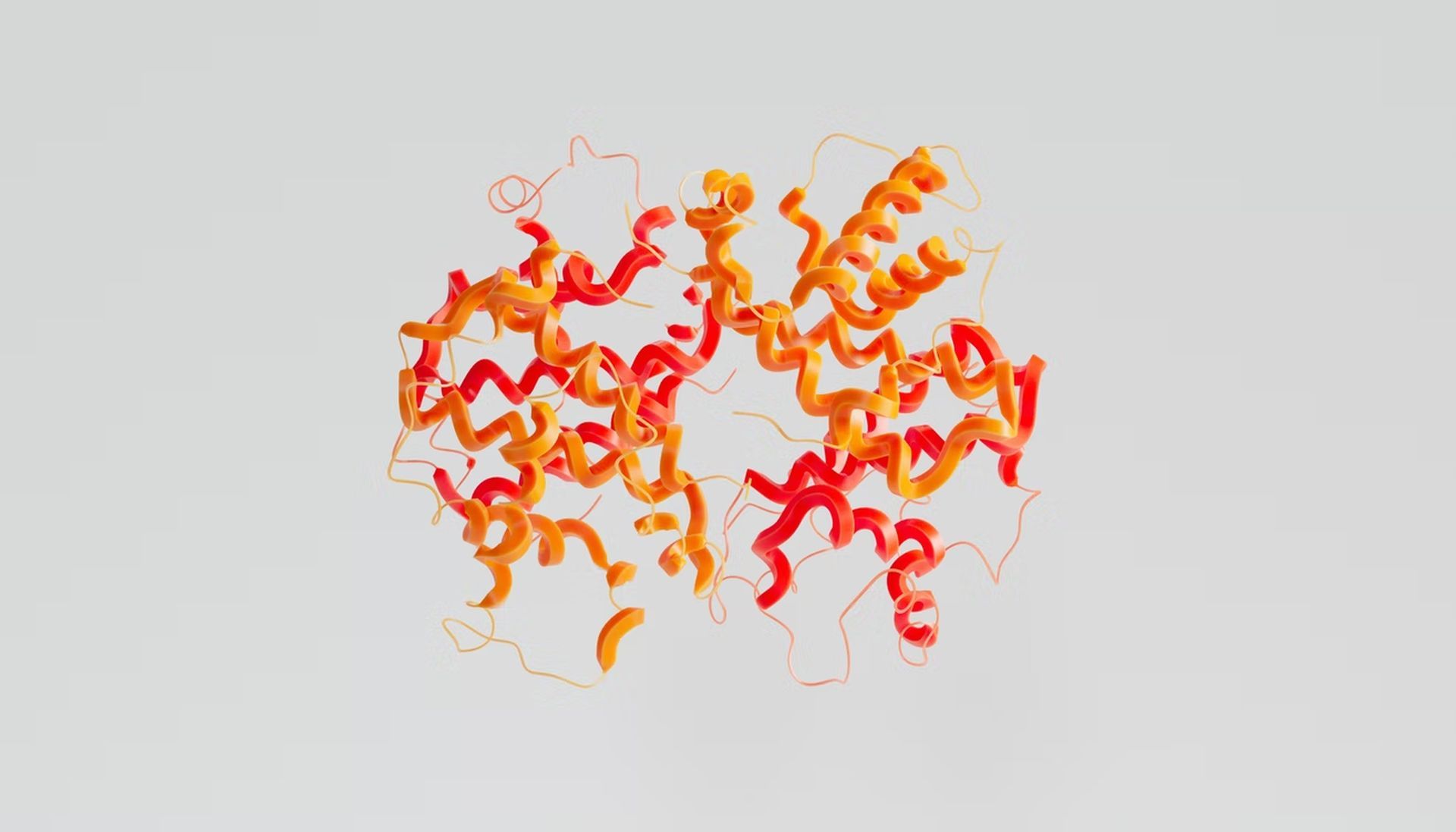
Each of these proteins adopts a distinct 3D structure in the body through folding the chains of amino acids that make up each one of them, which is crucial to how well they function.
Biologists have tried to predict the structures of proteins using costly experimental techniques, such as the tedious use of time-consuming techniques like X-ray crystallography or electron microscopy for decades.
Since the invention of computers, scientists have created virtual simulations of how the amino acid chains that makeup proteins would fold in various scenarios, resulting in the overall 3D structure of proteins.
Since AlphaFold’s introduction in 2020, more than 500,000 scientists have used it to decipher the structure of “virtually all listed proteins known to science.”
According to the business, AlphaFold learns to decode the remaining protein folding structures from around 100,000 previously known protein folding structures that scientists have already solved.

With the potential to speed up work on significant real-world issues “ranging from plastic pollution to antibiotic resistance,” the most recent development will increase the AlphaFold Protein Structure Database (AlphaFold DB) from approximately 1 million structures to over 200 million structures.
AI-based MARL method improved cooperation between teams of robots
The company stated that adding the anticipated structures for proteins found in plants, bacteria, animals, and other creatures in the new version may help with significant global problems, “including sustainability, food insecurity, and neglected diseases.”
“You can think of it as covering the entire protein universe. We’re at the beginning of a new era now in digital biology,” said Demis Hassabis, the Head of DeepMind.
With the help of the new structural predictions, researchers can determine whether or not disease-related protein variant forms exist.
For instance, AlphaFold protein structure predictions are assisting in the discovery of medications for neglected tropical diseases like leishmaniasis and Chagas disease, ailments that disproportionately afflict people in the world’s impoverished regions.
Additionally, Yale University researchers used the AlphaFold database in April to create a brand-new malaria vaccine.
The environmental impact of AI makes regulations vital for a sustainable future
Scientists can create medications that can successfully activate or play the role of malfunctioning proteins. They can also repress those creating issues by deciphering the structures of important proteins in the body associated with diseases.
Understanding protein structures can help treat diseases and be used to design solutions to worldwide environmental issues.

For instance, scientists and DeepMind‘s AI have collaborated to create faster-acting enzymes to degrade and recycle some of the most environmentally harmful single-use plastics.
“AlphaFold is the singular and momentous advance in life science that demonstrates the power of AI. Determining the 3D structure of a protein used to take many months or years; it now takes seconds,” Eric Topol, Director of the Scripps Research Translational Institute, said.
“AlphaFold has already accelerated and enabled massive discoveries, including cracking the structure of the nuclear pore complex. And with this new addition of structures illuminating nearly the entire protein universe, we can expect more biological mysteries to be solved each day,” he added.